Quantile Regression
Introduction
Quantile regression is another variation on least squares . The loss is the tilted \(l_1\) function,
\[ \phi(u) = \tau\max(u,0) - (1-\tau)\max(-u,0) = \frac{1}{2}|u| + \left(\tau - \frac{1}{2}\right)u, \]
where \(\tau \in (0,1)\) specifies the quantile. The problem as before
is to minimize the total residual loss. This model is commonly used in
ecology, healthcare, and other fields where the mean alone is not
enough to capture complex relationships between variables. CVXR
allows us to create a function to represent the loss and integrate it
seamlessly into the problem definition, as illustrated below.
Example
We will use an example from the quantreg package. The vignette
provides an example of the estimation and plot.
data(engel)
p <- ggplot(data = engel) +
geom_point(mapping = aes(x = income, y = foodexp), color = "blue")
taus <- c(0.1, 0.25, 0.5, 0.75, 0.90, 0.95)
fits <- data.frame(
coef(lm(foodexp ~ income, data = engel)),
sapply(taus, function(x) coef(rq(formula = foodexp ~ income, data = engel, tau = x))))
names(fits) <- c("OLS", sprintf("$\\tau_{%0.2f}$", taus))
nf <- ncol(fits)
colors <- colorRampPalette(colors = c("black", "red"))(nf)
p <- p + geom_abline(intercept = fits[1, 1], slope = fits[2, 1], color = colors[1], size = 1.5)## Warning: Using `size` aesthetic for lines was deprecated in ggplot2 3.4.0.
## ℹ Please use `linewidth` instead.
## This warning is displayed once every 8 hours.
## Call `lifecycle::last_lifecycle_warnings()` to see where this warning was generated.for (i in seq_len(nf)[-1]) {
p <- p + geom_abline(intercept = fits[1, i], slope = fits[2, i], color = colors[i])
}
p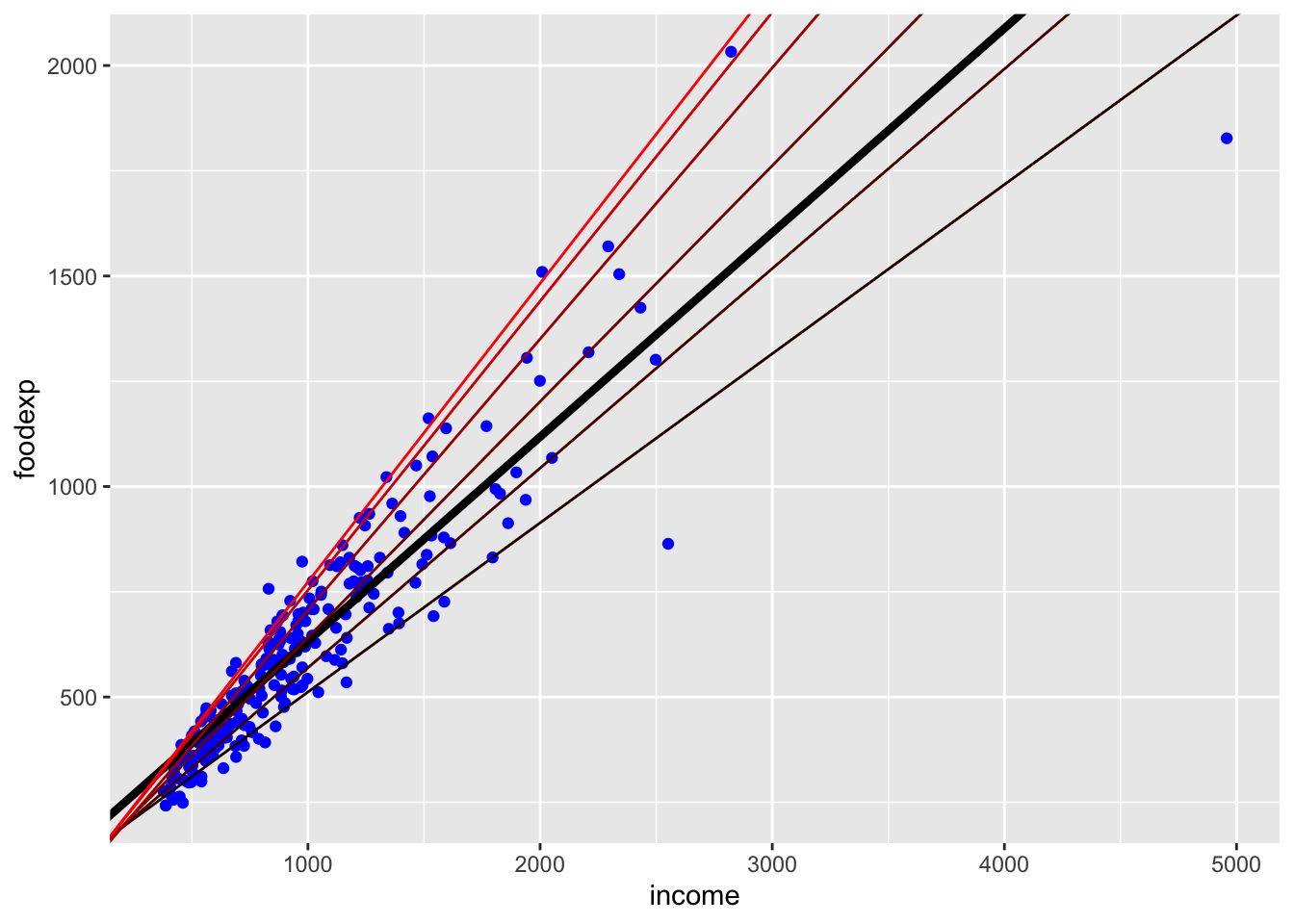
The above plot shows the quantile regression fits for \(\tau = (0.1, 0.25, 0.5, 0.75, 0.90, 0.95)\). The OLS fit is the thick black line.
The following is a table of the estimates.
knitr::kable(fits, format = "html", caption = "Fits from OLS and `quantreg`") %>%
kable_styling("striped") %>%
column_spec(1:8, background = "#ececec")| OLS | \(\tau_{0.10}\) | \(\tau_{0.25}\) | \(\tau_{0.50}\) | \(\tau_{0.75}\) | \(\tau_{0.90}\) | \(\tau_{0.95}\) | |
|---|---|---|---|---|---|---|---|
| (Intercept) | 147.4753885 | 110.1415742 | 95.4835396 | 81.4822474 | 62.3965855 | 67.3508721 | 64.1039632 |
| income | 0.4851784 | 0.4017658 | 0.4741032 | 0.5601806 | 0.6440141 | 0.6862995 | 0.7090685 |
The CVXR formulation follows. Note we make use of model.matrix to
get the intercept column painlessly.
X <- model.matrix(foodexp ~ income, data = engel)
y <- matrix(engel[, "foodexp"], ncol = 1)
beta <- Variable(2)
quant_loss <- function(u, tau) { 0.5 * abs(u) + (tau - 0.5) * u }
solutions <- sapply(taus, function(tau) {
obj <- sum(quant_loss(y - X %*% beta, tau = tau))
prob <- Problem(Minimize(obj))
## THE OSQP solver returns an error for tau = 0.5
solve(prob, solver = "ECOS")$getValue(beta)
})
fits <- data.frame(coef(lm(foodexp ~ income, data = engel)),
solutions)
names(fits) <- c("OLS", sprintf("$\\tau_{%0.2f}$", taus))Here is a table similar to the above with the OLS estimate added in for easy comparison.
knitr::kable(fits, format = "html", caption = "Fits from OLS and `CVXR`") %>%
kable_styling("striped") %>%
column_spec(1:8, background = "#ececec")| OLS | \(\tau_{0.10}\) | \(\tau_{0.25}\) | \(\tau_{0.50}\) | \(\tau_{0.75}\) | \(\tau_{0.90}\) | \(\tau_{0.95}\) | |
|---|---|---|---|---|---|---|---|
| (Intercept) | 147.4753885 | 110.1415744 | 95.4835373 | 81.4822486 | 62.3965854 | 67.3508709 | 64.1039682 |
| income | 0.4851784 | 0.4017658 | 0.4741032 | 0.5601805 | 0.6440141 | 0.6862995 | 0.7090685 |
The results match.
## Testthat Results: No output is goodSession Info
sessionInfo()## R version 4.4.1 (2024-06-14)
## Platform: x86_64-apple-darwin20
## Running under: macOS Sonoma 14.5
##
## Matrix products: default
## BLAS: /Library/Frameworks/R.framework/Versions/4.4-x86_64/Resources/lib/libRblas.0.dylib
## LAPACK: /Library/Frameworks/R.framework/Versions/4.4-x86_64/Resources/lib/libRlapack.dylib; LAPACK version 3.12.0
##
## locale:
## [1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
##
## time zone: America/Los_Angeles
## tzcode source: internal
##
## attached base packages:
## [1] stats graphics grDevices datasets utils methods base
##
## other attached packages:
## [1] quantreg_5.98 SparseM_1.84 kableExtra_1.4.0 ggplot2_3.5.1
## [5] CVXR_1.0-15 testthat_3.2.1.1 here_1.0.1
##
## loaded via a namespace (and not attached):
## [1] gtable_0.3.5 xfun_0.45 bslib_0.7.0 lattice_0.22-6
## [5] Rmosek_10.2.0 vctrs_0.6.5 tools_4.4.1 generics_0.1.3
## [9] tibble_3.2.1 fansi_1.0.6 highr_0.11 pkgconfig_2.0.3
## [13] Matrix_1.7-0 desc_1.4.3 assertthat_0.2.1 lifecycle_1.0.4
## [17] compiler_4.4.1 farver_2.1.2 stringr_1.5.1 MatrixModels_0.5-3
## [21] brio_1.1.5 munsell_0.5.1 gurobi_11.0-0 codetools_0.2-20
## [25] ECOSolveR_0.5.5 htmltools_0.5.8.1 sass_0.4.9 cccp_0.3-1
## [29] yaml_2.3.9 gmp_0.7-4 pillar_1.9.0 crayon_1.5.3
## [33] jquerylib_0.1.4 MASS_7.3-61 rcbc_0.1.0.9001 clarabel_0.9.0
## [37] Rcplex_0.3-6 cachem_1.1.0 tidyselect_1.2.1 digest_0.6.36
## [41] slam_0.1-50 stringi_1.8.4 dplyr_1.1.4 bookdown_0.40
## [45] labeling_0.4.3 splines_4.4.1 rprojroot_2.0.4 fastmap_1.2.0
## [49] grid_4.4.1 colorspace_2.1-0 cli_3.6.3 magrittr_2.0.3
## [53] survival_3.7-0 utf8_1.2.4 withr_3.0.0 Rmpfr_0.9-5
## [57] scales_1.3.0 bit64_4.0.5 rmarkdown_2.27 bit_4.0.5
## [61] blogdown_1.19 evaluate_0.24.0 knitr_1.48 Rglpk_0.6-5.1
## [65] viridisLite_0.4.2 rlang_1.1.4 Rcpp_1.0.12 glue_1.7.0
## [69] xml2_1.3.6 pkgload_1.4.0 svglite_2.1.3 rstudioapi_0.16.0
## [73] jsonlite_1.8.8 R6_2.5.1 systemfonts_1.1.0