Sparse Inverse Covariance Estimation
Introduction
Assume we are given i.i.d. observations xi∼N(0,Σ) for i=1,…,m, and the covariance matrix Σ∈Sn+, the set of symmetric positive semidefinite matrices, has a sparse inverse S=Σ−1. Let Q=1m−1∑mi=1(xi−ˉx)(xi−ˉx)T be our sample covariance. One way to estimate Σ is to maximize the log-likelihood with the prior knowledge that S is sparse (Friedman, Hastie, and Tibshirani 2008), which amounts to the optimization problem:
maximizeSlogdet
The parameter \alpha \geq 0 controls the degree of sparsity. The problem
is convex, so we can solve it using CVXR.
Example
We’ll create a sparse positive semi-definite matrix S using synthetic data
set.seed(1)
n <- 10 ## Dimension of matrix
m <- 1000 ## Number of samples
## Create sparse, symmetric PSD matrix S
A <- rsparsematrix(n, n, 0.15, rand.x = stats::rnorm)
Strue <- A %*% t(A) + 0.05 * diag(rep(1, n)) ## Force matrix to be strictly positive definiteWe can now create the covariance matrix R as the inverse of S.
R <- base::solve(Strue)As test data, we sample from a multivariate normal with the fact that if Y \sim N(0, I), then R^{1/2}Y \sim N(0, R) since R is symmetric.
x_sample <- matrix(stats::rnorm(n * m), nrow = m, ncol = n) %*% t(expm::sqrtm(R))
Q <- cov(x_sample) ## Sample covariance matrixFinally, we solve our convex program for a set of \alpha values.
Version 1.0 Note: Positive semi-definite variables are now
designated using PSD = TRUE rather than the Semidef function!
alphas <- c(10, 8, 6, 4, 1)
if (packageVersion("CVXR") > "0.99-7") {
S <- Variable(n, n, PSD = TRUE)
} else {
S <- Semidef(n) ## Variable constrained to positive semidefinite cone
}
obj <- Maximize(log_det(S) - matrix_trace(S %*% Q))
S.est <- lapply(alphas,
function(alpha) {
constraints <- list(sum(abs(S)) <= alpha)
## Form and solve optimization problem
prob <- Problem(obj, constraints)
result <- solve(prob)
## Create covariance matrix
R_hat <- base::solve(result$getValue(S))
Sres <- result$getValue(S)
Sres[abs(Sres) <= 1e-4] <- 0
Sres
})In the code above, the Semidef constructor restricts S to the
positive semidefinite cone. In our objective, we use CVXR functions
for the log-determinant and trace. The expression matrix_trace(S %*% Q) is equivalent to `sum(diag(S %*% Q))}, but the former is preferred
because it is more efficient than making nested function calls.
However, a standalone atom does not exist for the determinant, so we
cannot replace log_det(S) with log(det(S)) since det is
undefined for a Semidef object.
## Testthat Results: No output is goodResults
The figures below depict the solutions for the above dataset with m = 1000, n = 10, and S containing 26% non-zero entries, represented by the dark squares in the images below. The sparsity of our inverse covariance estimate decreases for higher \alpha, so that when \alpha = 1, most of the off-diagonal entries are zero, while if \alpha = 10, over half the matrix is dense. At \alpha = 4, we achieve the true percentage of non-zeros.
do.call(multiplot, args = c(list(plotSpMat(Strue)),
mapply(plotSpMat, S.est, alphas, SIMPLIFY = FALSE),
list(layout = matrix(1:6, nrow = 2, byrow = TRUE))))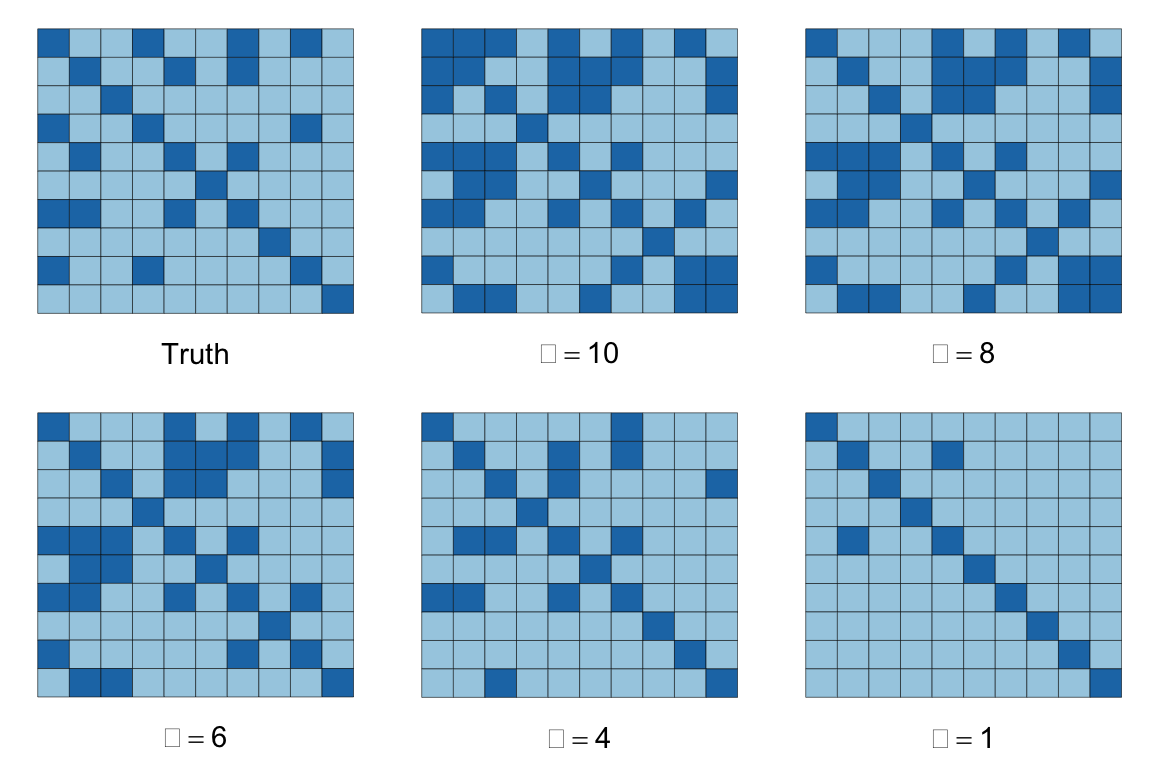
Session Info
sessionInfo()## R version 4.4.2 (2024-10-31)
## Platform: x86_64-apple-darwin20
## Running under: macOS Sequoia 15.1
##
## Matrix products: default
## BLAS: /Library/Frameworks/R.framework/Versions/4.4-x86_64/Resources/lib/libRblas.0.dylib
## LAPACK: /Library/Frameworks/R.framework/Versions/4.4-x86_64/Resources/lib/libRlapack.dylib; LAPACK version 3.12.0
##
## locale:
## [1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
##
## time zone: America/Los_Angeles
## tzcode source: internal
##
## attached base packages:
## [1] grid stats graphics grDevices datasets utils methods
## [8] base
##
## other attached packages:
## [1] expm_1.0-0 Matrix_1.7-1 ggplot2_3.5.1 CVXR_1.0-15
## [5] testthat_3.2.1.1 here_1.0.1
##
## loaded via a namespace (and not attached):
## [1] gmp_0.7-5 clarabel_0.9.0.1 sass_0.4.9 utf8_1.2.4
## [5] generics_0.1.3 slam_0.1-54 blogdown_1.19 lattice_0.22-6
## [9] digest_0.6.37 magrittr_2.0.3 RColorBrewer_1.1-3 evaluate_1.0.1
## [13] bookdown_0.41 pkgload_1.4.0 fastmap_1.2.0 rprojroot_2.0.4
## [17] jsonlite_1.8.9 brio_1.1.5 scs_3.2.4 Rmosek_10.2.0
## [21] fansi_1.0.6 scales_1.3.0 codetools_0.2-20 jquerylib_0.1.4
## [25] cli_3.6.3 Rmpfr_0.9-5 rlang_1.1.4 Rglpk_0.6-5.1
## [29] bit64_4.5.2 munsell_0.5.1 withr_3.0.2 cachem_1.1.0
## [33] yaml_2.3.10 tools_4.4.2 Rcplex_0.3-6 rcbc_0.1.0.9001
## [37] dplyr_1.1.4 colorspace_2.1-1 gurobi_11.0-0 assertthat_0.2.1
## [41] vctrs_0.6.5 R6_2.5.1 lifecycle_1.0.4 bit_4.5.0
## [45] desc_1.4.3 cccp_0.3-1 pkgconfig_2.0.3 bslib_0.8.0
## [49] pillar_1.9.0 gtable_0.3.6 glue_1.8.0 Rcpp_1.0.13-1
## [53] highr_0.11 xfun_0.49 tibble_3.2.1 tidyselect_1.2.1
## [57] knitr_1.48 farver_2.1.2 htmltools_0.5.8.1 labeling_0.4.3
## [61] rmarkdown_2.29 compiler_4.4.2